Justification of Toxin Assay Samples
Deciding on the number and which samples to be assessed for C. difficile toxin levels.
Justification of Toxin Assay Samples
Criteria:
- Want at least 2 biological replicates
- Want Days 1 and 10. As well as other days common to most samples.
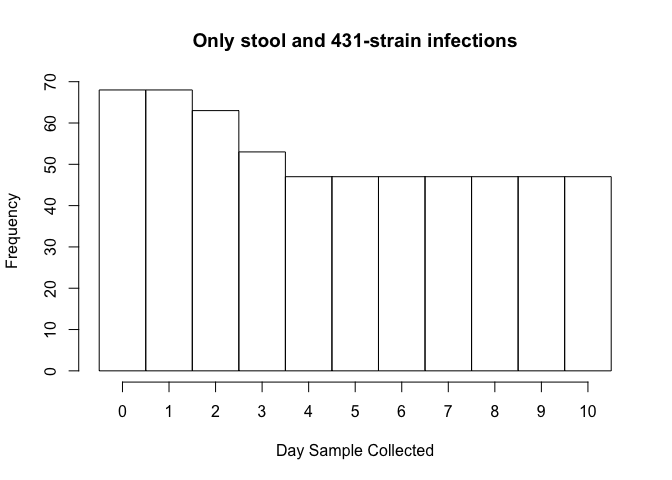
The histogram below shows the frequency of samples by day.
metadata <- read.table(file = "~/Documents/Schloss/Data/GF/GFmetadata.txt",
header = T, na.strings = "NA", stringsAsFactors = FALSE)
hist.info <- hist(metadata[metadata$sample_type == "stool" & metadata$cdiff_strain ==
"431" & !is.na(metadata$day), "day"], breaks = c(seq(-1, 10)), xlab = "Day Sample Collected",
xaxt = "n", main = "Only stool and 431-strain infections")
axis(side = 1, at = hist.info$mids, labels = seq(0, 10))
# Number of groups with Day 1, 2, 3, 4, 7, 10 samples
length(unique(metadata[metadata$sample_type == "stool" & metadata$cdiff_strain ==
"431" & !is.na(metadata$day) & metadata$day == "1", "cage_id"]))
## [1] 22
length(unique(metadata[metadata$sample_type == "stool" & metadata$cdiff_strain ==
"431" & !is.na(metadata$day) & metadata$day == "2", "cage_id"]))
## [1] 21
length(unique(metadata[metadata$sample_type == "stool" & metadata$cdiff_strain ==
"431" & !is.na(metadata$day) & metadata$day == "3", "cage_id"]))
## [1] 17
length(unique(metadata[metadata$sample_type == "stool" & metadata$cdiff_strain ==
"431" & !is.na(metadata$day) & metadata$day == "4", "cage_id"]))
## [1] 15
length(unique(metadata[metadata$sample_type == "stool" & metadata$cdiff_strain ==
"431" & !is.na(metadata$day) & metadata$day == "7", "cage_id"]))
## [1] 15
length(unique(metadata[metadata$sample_type == "stool" & metadata$cdiff_strain ==
"431" & !is.na(metadata$day) & metadata$day == "10", "cage_id"]))
## [1] 15
Day 2 and 3 are common to many samples and are the ending points for several groups. These days should be processed. I’m considering additionally either doing day 4 OR doing a day towards the middle of 3-10, i.e. either 6 or 7. Depending on the total number of samples this brings me to, possibly days 4 and 7.
Update
After estimating the number of samples (using 2 or 3 mice per group) and laying out the first plate, I’ve realized that we have about 100 less samples than Jhansi originally planned for (~400 samples). Arraying the first plate, I observed that while I can plan each mouse I want to be processed, there isn’t always enough sample. So we may just move forward with doing every sample available on the days selected for each group. Majority of these are stool supernatant samples; however there are some groups, particularly the severe CDI cases, where only cecal content could be collected. Koenigsknecht MJ et al. 2015 showed that at 24hrs post infection (using the cefoperazone CDI model), the levels of toxin in the cecum and stool were fairly similar, differing by less than a log (log TOPS). I’m using what’s available to me; so we are going to go ahead and look at the toxin in the cecal content of mice if stool is unavailable. The days we are assessing toxin are days 1, 2, 3, 4, 7, and 10. I estimate the max total number of samples to be around 330 for these selected days.